RNA Library

The recent substantial progress in RNA biology underscores the importance of RNA in normal and aberrant cellular functions. RNA is essential for transcriptional regulation, translational regulation, protein function, and catalysis, responsibilities that have classically been reserved for proteins. It also highlights the potential of targeting RNA for treatment of a multitude of diseases including bacterial/viral infection and cancer.
Library Design
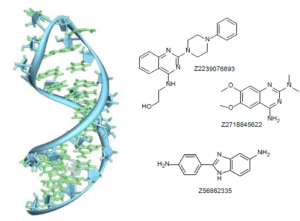
RNAs can form well-defined secondary structures, such as double helices, hairpins, bulges, internal loop, stems, which offer a structural basis for designing therapeutic agents. These structural features have been taken into account in the design of our RNA Targeted Library. We focused on compounds which bind to RNA, forming interactions with different types of secondary structures.
Reference Set of active compounds has been collected from public sources/databases (ChEMBL, PubChem, Drugs.com and other), then filtered to remove non-selective binders and compounds with reactive moieties. Similarity search (chemical fingerprints) was carried out against MedChem filtered Enamine in-stock collection, 2.4 M compounds, to yield a subset of 400 molecules.
Pharmacophore search and Shape similarity search were used to enrich the library with new structures that can form the same types of interaction as known ligand. Three 3D pharmacophore models were used for in silico screening based on the structure of most known binders. Additionally, the substructure search was applied to find molecules bearing common structural moieties and cores known to be important for interaction with RNA (Inforna database was used for substructure selection).
The library comprises only drug-like compounds and provides an excellent basis for posttranscriptional gene regulation researches, antiviral and antibacterial drug discovery projects.